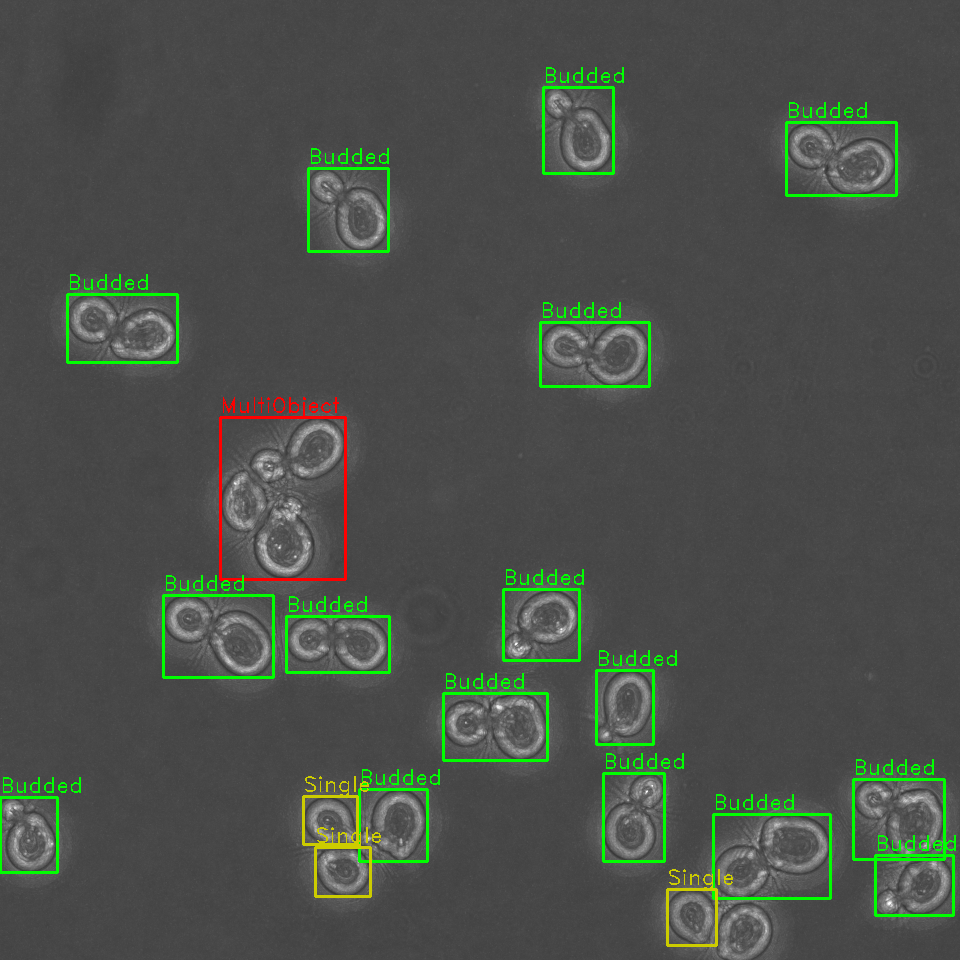
Rapid automated identification, classification, and cartography of yeast cells for shape analysis allows for sorting of cells by budding cycle stage and better downstream segmentation.
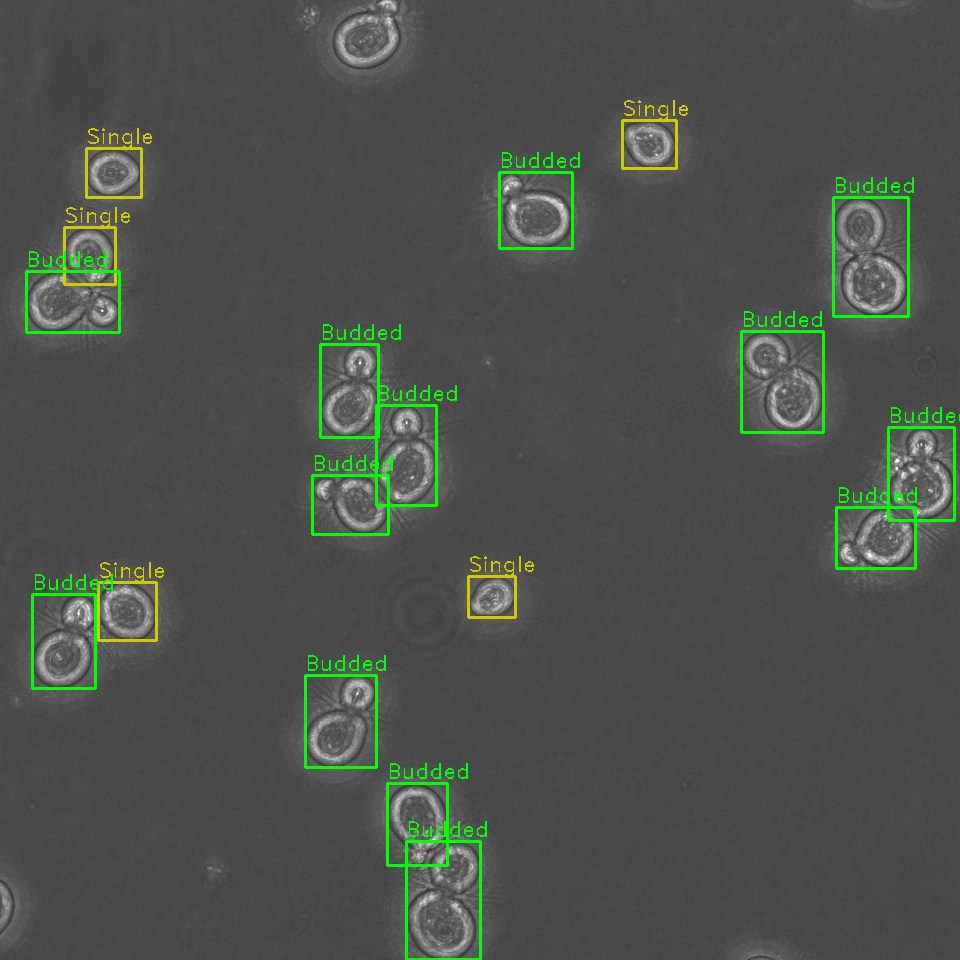
Identify, classify, and sort cells within a field of view by budding stage
Identification and sorting of individual cells from a microscopy image permits for:
- Tracking of cell budding stage as either single or budded (along with approximate stage of budding).
- Characterization of the distribution of various cell morphologies within a population
- Identification of dead cells to omit from downstream analyses
Identify problematic cell clusters for further analysis
Additionally, our algorithm is capable of detecting clusters of cells, where it is difficult to determine the boundaries of multiple cells in close proximity. These cells are annotated and can be opted out from future analyses, or later manually examined.
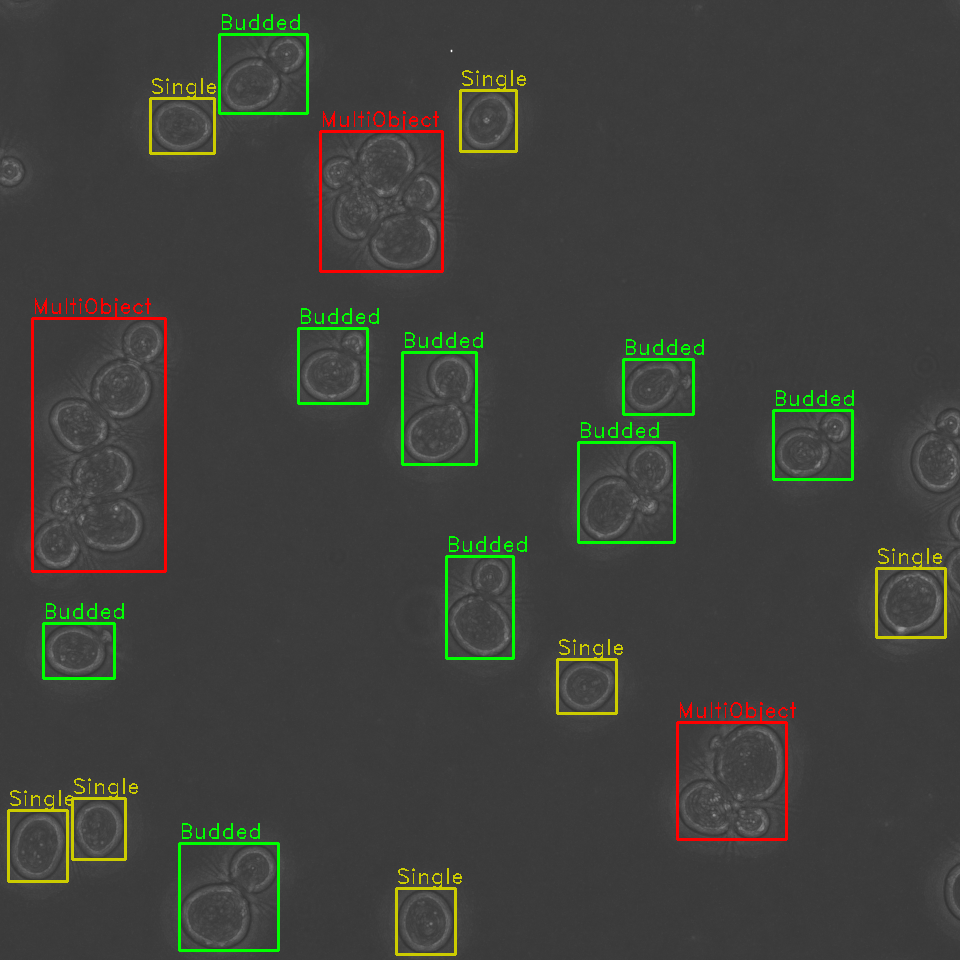
When run in tandem with other segmentation pipelines, pre-isolation of individual cells from a larger image allows for more robust fluorescence segmentation pipelines. One of the common problems with binary segmentation of a fluorescent marker is cell-to-cell heterogeneity in marker expression or uptake. Isolation of individual cells allows for more accurate local thresholding for fluorescence segmentation and increases the workable signal to noise ratio range for detection of low-fluorescence objects.