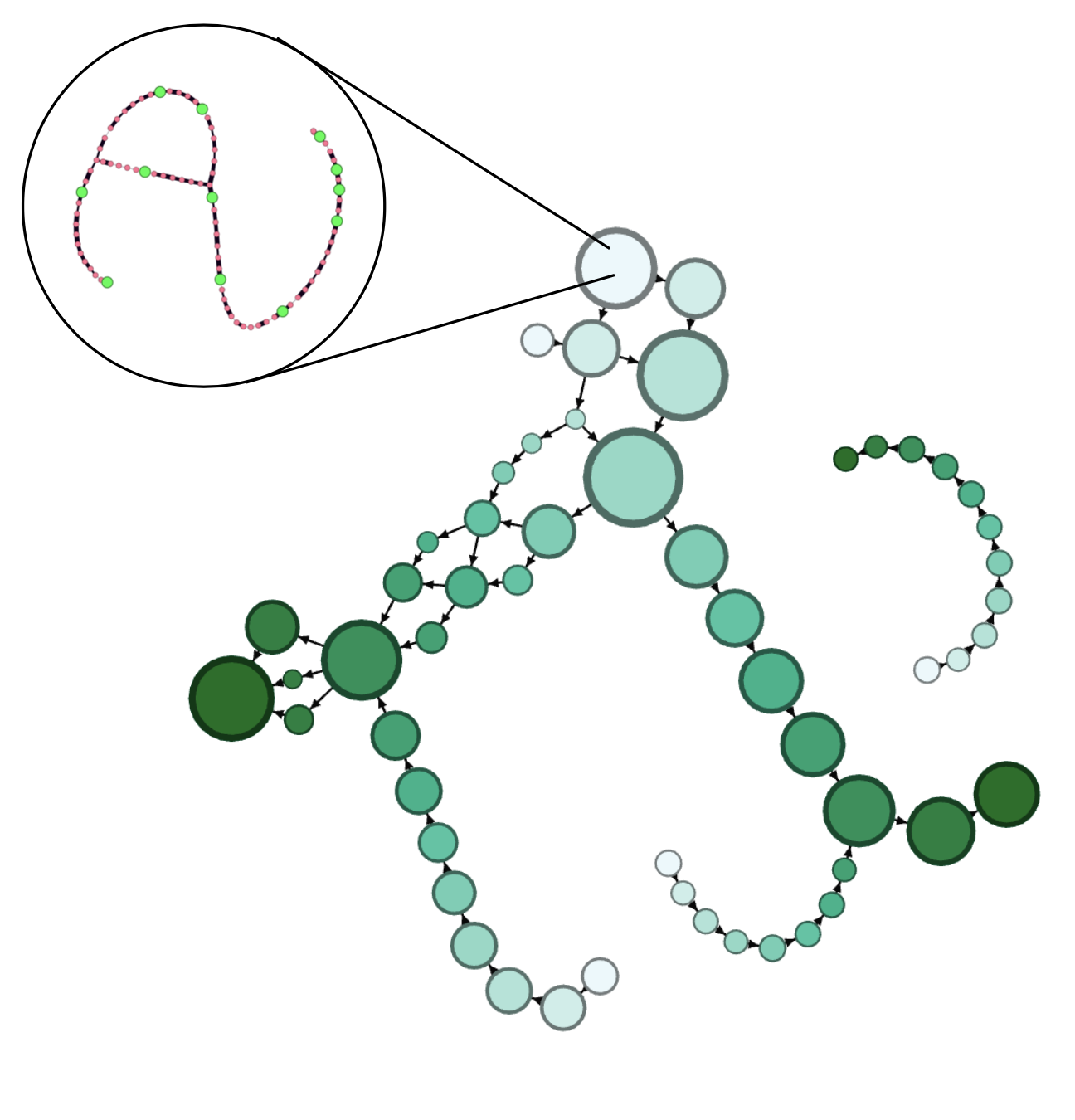
Runs common network analysis algorithms and structure-to-structure analysis on skeletonization of mitochondrial networks,
Analyze surface-to-surface statistics with respect to structure
One of the unique aspects of mitochondria structure is its ability to form dynamic reticulated structures. Tracking of objects relative to that underlying structure is difficult to do by hand, as most multidimensional image analysis programs do not run backbone tracing algorithms or feature detection. Our pipeline enables characterizing localization of structures with respect to other features of other structures, where a user can examine for instance how far away one organelle is to a particular feature of another organelle.
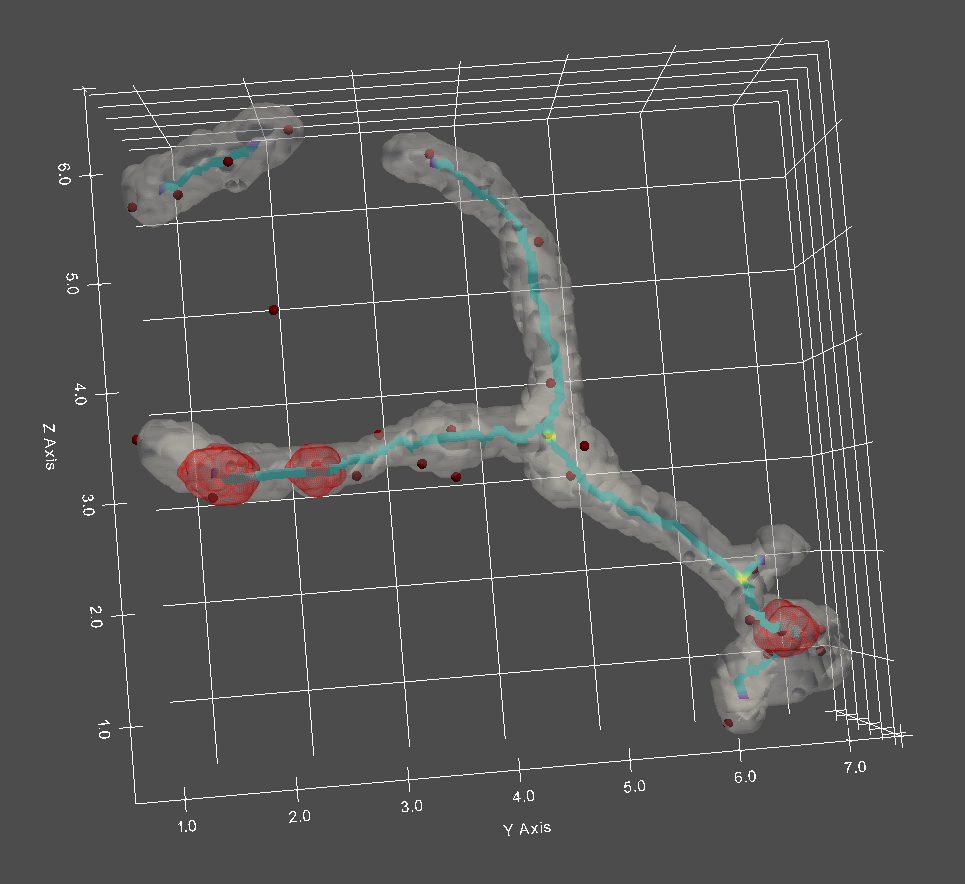
In our case, with mitochondria we can track positioning of other organelles with respect to underlying mitochondrial network structure. With static images, these analyses would include (and are not limited to) positioning of punctate and/or large organelle structures (such as nucleoids, fission machinery, ER-Mito contact sites) with respect to mitochondria branch, tip, or hub points. The underlying network wireframe structure in the sample rendering below has also been color coded as to the degree of the network structure (branch, hub, tip).
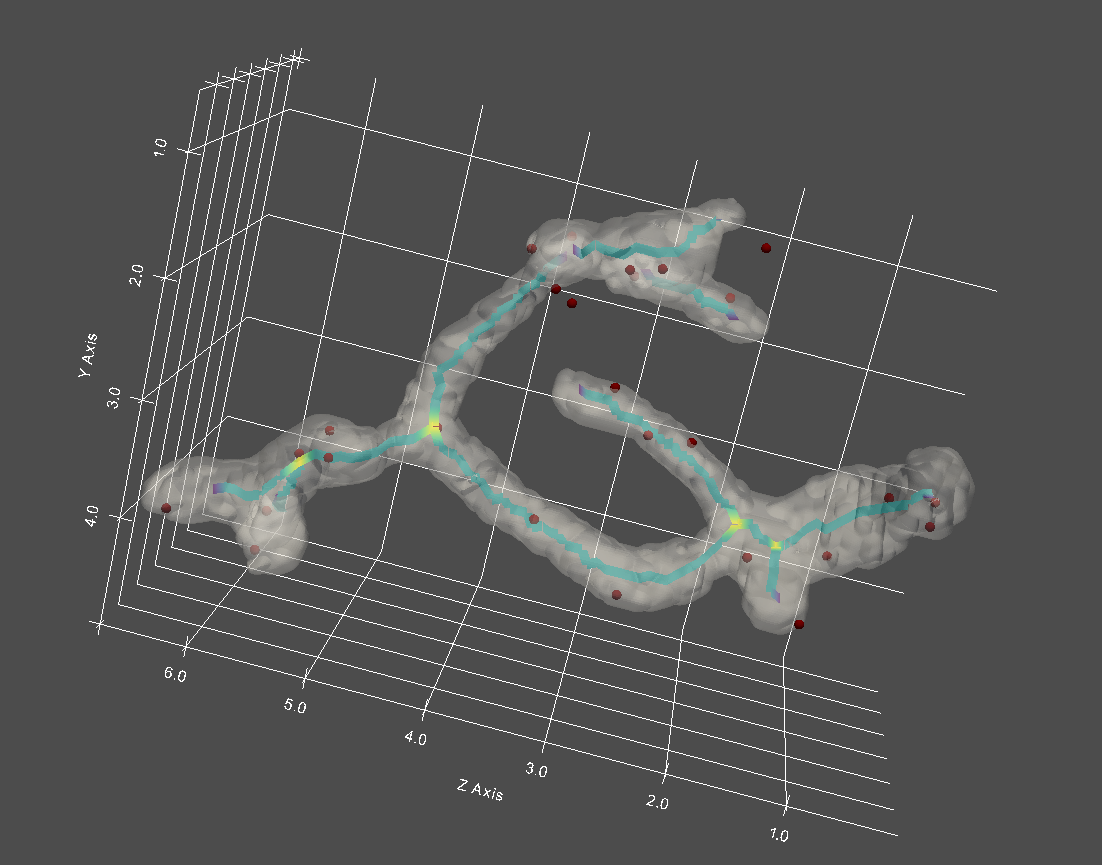
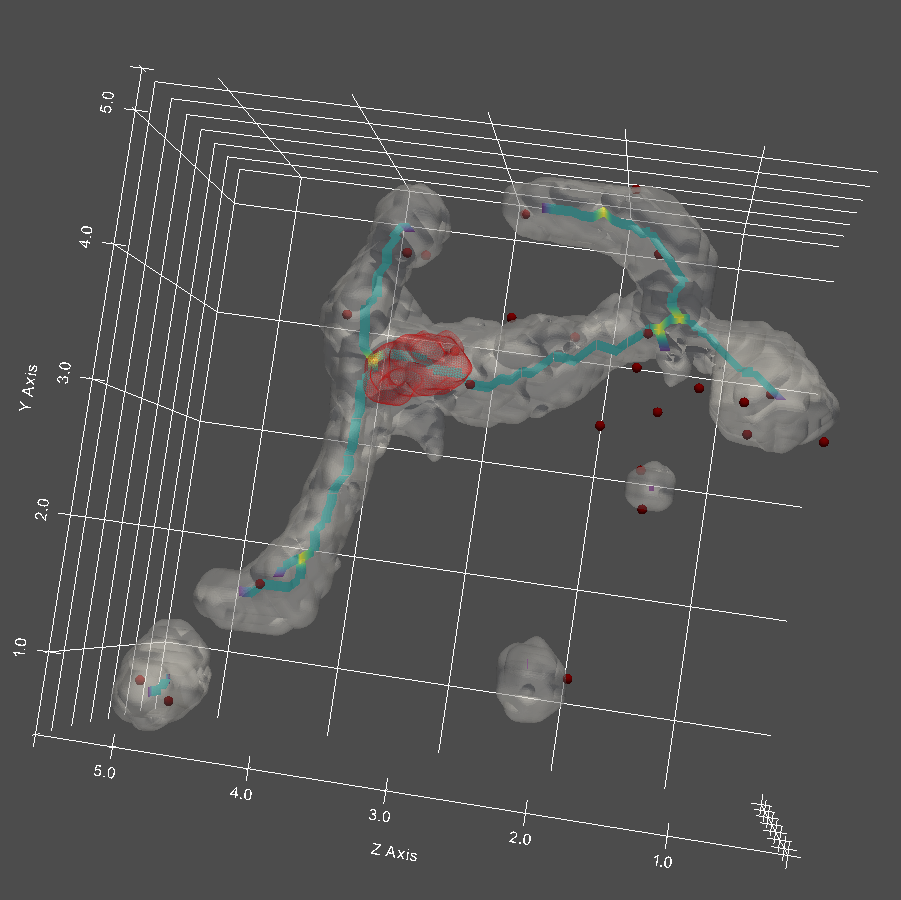
Run queries on mitochondria substructure and event dynamics
Mitochondria are furthermore dynamic structures, and it may be useful to analyze the evolution of structure-structure localization/proximity over time in microscopy images. With our pipeline, not only can we track the evolution of the mitochondria network over time, but also how positioning of mitochondria sub-structures (such as nucleoids) evolves over time.
Our tracking is furthermore not only limited to structures - we can track position bias of various network remodeling events. Mitochondria undergo fission and fusion, and the placement of fission and/or fusion sites has a role on the underlying structure of the mitochondria network. Our analysis pipeline is capable of characterizing positioning these remodeling events with respect to the mitochondria network and/or any associated structures (i.e. proximity of ERMES, nucleoids, to fission/fusion sites).
Run location specific network analysis
Using our cellular localization pipeline in tandem, this pipeline can extract statistics on how bud vs the mother cell mitochondria are differently structured, undergo different remodeling dynamics, and much more. Existing analyses in this pipeline can be further focused with respect to organelle localization within the mother-bud system.